MD simulation of proteins immobilized onto gold substrates
MD simulation of plastocyanin, adsorbed onto a gold surface has been performed starting from the crystallographic structure of a poplar plastocyanin mutant engineered with the insertion of a disulfide bridge. The protein has been anchored to a gold substrate modelled by a cluster of three layers in the Au<111> configuration.
An analysis of structural and dynamical properties of the protein molecule, covalently anchored to the gold surface through either one (PCSS-I) or two (PCSS-II) sulfur atoms has been performed and compared with those of the free protein.
These results can provide some hints to interpret both topological experimental data as obtained by Atomic Force Microscopy (AFM) and ET properties investigated by Scanning Tunnelling Microscopy (STM).
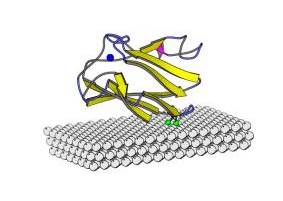
FIGURE: Model of the mutated plastocyanin anchored onto Au(111) surface by covalently binding of both the protein sulfurs to the gold substrate. Each sulfur is bound to three gold atoms. The sulfur atoms involved into the protein-gold interactions and the copper atom with its ligands are also shown.
The orientation and precession of the protein macromolecule about the normal to the gold surface has been investigated during the MD simulated trajectories for both the differently anchored systems.
An analysis of the temporal evolution of the axis joining the active site copper with a sulfur involved onto the protein-gold anchorage shows a different orientation and arrangement with respect to the gold surface of the protein systems anchored by one or two covalent bonds.
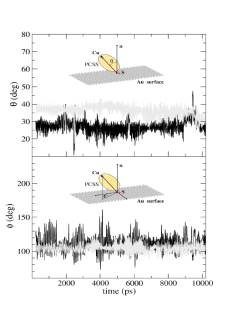
FIGURE: Temporal evolution, along a 10 ns MD trajectory, of the q (a) and j (b) angles describing the orientation and the precession, respectively, of the protein p axis with respect to gold surface, for the PCSS-I (dark gray) and PCSS-II (light gray). The average value and the standard deviation of q are (27 ± 4)° and (36 ± 3)° for PCSS-I and PCSS-II, respectively. The average values and the standard deviations of j are (112±12)° and (105 ± 6)° for PCSS-I and PCSS-II, respectively.
Inset: (a) q gives the angle between the p axis connecting the copper atom and the sulfur of Cys25 with the normal to the gold surface; (b) j gives the precession of this axis about the normal to the gold surface.
Back to Molecular Dynamics Simulation and Modelling