Protein docking and modelling
Protein docking is a computational method that aims to predict the
structure of a protein-protein complex starting from the crystallographic
coordinates of the single components by performing an exhaustive search of all
possible configurations of the two partners.
Azurin-cytocrome C551 complex
Computational methods have been used to model the interaction between Azurin,
an electron transfer protein from the opportunistic bacterium Pseudomonas aeruginosa, and its partner cytC551;
an experimental investigation being made difficult by the transient character
of the complex.
(
Azurin-p53
complex
In the last decade
it has been found that the bacterial protein Azurin is able to penetrate
cancerous cells and inhibits their uncontrolled proliferation. Surprisingly the
Azurin anticancer activity is connected with its interaction with the well
known tumour suppressor p53 which should be stabilized and reactivated in
cancer cells just by azurin. In this context, we have used computational
docking to model the interaction of Azurin with p53 domains, in order to
elucidate the molecular details of the interaction connected with Azurin mode
of action.
De Grandis V, Bizzarri AR,
Cannistraro S. Docking study
and
free energy simulation of the complex between
p53
DNA-binding domain and azurin. J. Mol. Recognit. 2007; 20: 215–226
Taranta M,
between
the N-terminal domain of the tumor suppressor
p53 and azurin. J. Mol. Recognit. 2009; 22: 215–222
Computational mutagenesis
Computational
methods allow simulating the effects that point mutations could have on the
structure and stability of a complex. By means of this procedure we have demonstrate
the crucial role of two Azurin amino acidic residues for its interaction with
p53. In such a way we have thus significantly contributed in the understanding
of the Azurin/p53 complex structure and properties.
From Azurin to p28
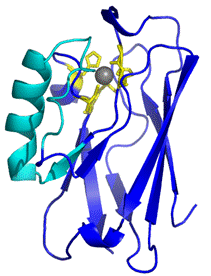
Three dimensional structure of Azurin. The cyan sequence corresponds to
p28.
In the last
two years the equip of scientist of the Division
of Surgical Oncology of the University of Illinois College of Medicine of
Chicago has found that the peptide fragment
of azurin called p28, (residues 50 - 77 of the whole protein), retains both the
cellular penetration ability and antiproliferative action of the whole protein
(Yamada et al., 2009). As for azurin, the p28 antiproliferative activity is consequent
to its interaction with p53.
Even if p28
has already entered the phase II clinical trials under the FDA allowance its mode
of action has not been clarified mostly because the details of its interaction
with p53 are unknown.
By means of computational docking procedure we have defined the
molecular details of the interaction of p28 with the p53 core domain
thus contributing
to open new perspectives on the possible p28 mode of action.
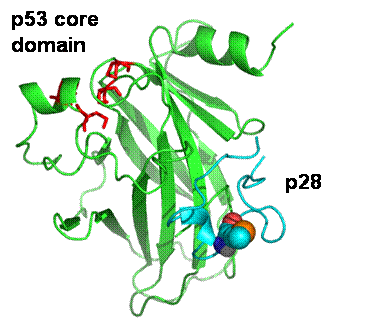
Three dimensional structure of the best p28-p23 core domain complex resulting
from docking and molecular dynamics simulation procedures. (Santini
S,