Protein Engineering and Modelling
Our group is particularly focused in studying the structural and functional properties of copper proteins (i.e. azurin and plastocyanin) small proteins involved in the electron transport chains of bacteria and plants. Recently we have deepened (Ultramicroscopy, 2001; Surface Science, 2003) the study of their structure and function at single molecule level by Scanning Probe Microscopy (STM/ AFM). In this respect, disulphide or thiol groups, which can form a covalent interaction with gold, can be usefully exploited for a stable and oriented immobilisation of the molecules.
In order to engineer such groups we have firstly designed mutants by exploiting Modelling software, such as What If and S-S bond, helpful tools for the identification of sites where could be possible to introduce mutations keeping intact the native structure of the protein.
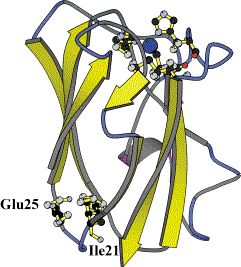
Subsequently we have applied site-directed mutagenesis to plastocyanin obtaining two mutants (in collaboration with Leiden University Metallo-Protein Group, headed by Prof. G.W. Canters and with Dr. A.G. Ficca). In the first one (PCSS), a disulphide bridge was inserted within the protein by selecting and modifying the residues Ile-21 to Cys and Glu-25 to Cys. In the second one (PCSH) a residue tail (threonine, cysteine and glycine) at the C-terminal position was inserted, see figure.
|
|
|
|
|
|
The integrity and functionality of metalloproteins can be assessed by using spectroscopic techniques, i.e. Resonance Raman and Electron Paramagnetic Resonance spectroscopy.
The mutant plastocyanin bearing the disulphide bridge (PCSS) shows spectroscopic features similar to the wild type plastocyanin (Arch. Biochem. Biophys., 2002). In the case of PCSH UV-Vis spectroscopy shows already that the tetrahedral geometry of the copper site is retained, although a further characterisation is still undergoing.
A close analysis of the three-dimensional macromolecular structure of the PCSS, achieved by crystallography (in collaboration with Prof. M. Bolognesi group in Genova) pointed out that the main three dimensional structure of the protein was unperturbed, as shown in the figure, and that the disulphide bound was established as shown in the comparison below between the wild-type (blue) and mutant (yellow) 21 and 25 loops.
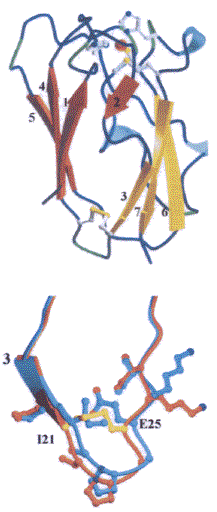
We are also planning a further structural analysis by NMR methods using INFM facilities at Roma Tor Vergata University in collaboration with Dr. F. Porcelli. These techniques are useful complements to the biochemical and functional characterisation of the proteins.